Real-Time PCR (qPCR): A Comprehensive Overview

A real-time polymerase chain reaction (PCR, or qPCR) is a molecular biology laboratory technique based on the polymerase chain reaction (PCR). It monitors the amplification of a specific DNA molecule during the PCR process rather than at the end, as in conventional PCR. Real-time PCR theoretically amplifies DNA exponentially, doubling the number of target molecules with each amplification cycle. Scientists reasoned when it was first developed that the number of cycles performed and the amount of PCR end-product could be used to compute the initial amount of DNA by comparing the material to a known standard. The technique of real-time quantitative PCR was developed in response to the need for robust quantification. At the moment, endpoint PCR is primarily used to amplify specific DNA for sequencing, cloning, and several other applications in other molecular biology techniques.
This section provides an introduction to real-time PCR, the benefits of Real-Time PCR, the four qPCR process steps, what real-time PCR is, and what products are available.
What is real-time PCR?
Real-time PCR, also known as quantitative or qPCR, measures the amount of PCR product present during a given cycle. The amplified DNA product, or amplicon, is detected in an endpoint analysis in conventional PCR. The accumulation of amplification products is measured in real-time as the reaction progresses, with product quantification after each cycle in real-time PCR.
Non-specific fluorescent dyes that intercalate with any double-stranded DNA and sequence-specific DNA probes consisting of oligonucleotides labelled with a fluorescent reporter that allows detection only after hybridisation of the probe with its complementary sequence are two common methods for detecting PCR products in real-time PCR.
PCR vs qPCR
| PCR | qPCR |
| PCR stands for polymerase chain reaction. | qPCR stands for quantitative polymerase chain reaction |
| PCR is a qualitative method of DNA amplification. However, 'Semi-quantitative' results can be obtained by comparing the intensity of the PCR-amplified sample on an agarose gel to a standard. | qPCR is a quantitative method. |
| PCR employs the enzyme DNA polymerase, which directs the synthesis of DNA on a single-strand DNA template from deoxynucleotide substrates. | The same PCR amplification principle is used in real-time PCR. However, rather than inspecting bands on a gel at the end of the PCR reaction, the PCR amplification products are measured in "real-time" with a detector or camera at the end of each cycle. |
| PCR has poor precision and low sensitivity. | With increased precision in a qPCR experiment, you can distinguish smaller differences in copy numbers of nucleic acids. |
| The data collected at the end of the reaction and thus requires post PCR processing. | The data is collected during the reaction's exponential phase. Thus, no post PCR processing is required. |
| PCR has a low resolution. | qPCR has a high resolution. |
The advantages of real-time PCR
The advantages of real-time PCR include:
- Real-time PCR is particularly useful in large commercial laboratories that process a large number of similar samples.
- Real-time detection systems are typically automated, and they can overcome some of the limitations of traditional PCR.
- qPCR detects amplified fragments during the amplification process.
- Measuring during the exponential phase is more effective because measurements taken during the plateau phase do not always clearly indicate the quantity of starting material. The initial number of copies of template DNA can be determined using real-time PCR.
The process steps of RT-PCR
The reverse transcriptase PCR procedure consists of steps which are given below:
Extraction:
RNA extraction is the first and foremost step of RT-qPCR.
Cell lysis is the first step in the RNA extraction process. A buffer containing Guanidine Thiocyanate or other chaotropes linearize the RNA polymer, disrupt hydrogen bonding, and render any RNAses present in the cell lysate inactive. They lyse cells by disbanding the phospholipid bilayer due to their ability to disrupt hydrogen bonding.
A homogenization column can help with homogenization. Viscous polymers such as DNA and lipids can be separated, and the cell lysate can flow more easily by passing at high speeds through resins.
Ethanol is then added to the sample to reduce the overall water concentration and precipitate proteins. RNA is highly soluble in water, as one would expect. The RNA from the cell lysate is then bound using a spin column.
A low chaotrope concentration wash buffer is initially used to clean the RNA sample and remove proteins while the RNA remains attached to the column. Following that, an ethanol wash removes some of the chaotropic salts left over from previous washes.
Finally, in the absence of chaotropic salts, RNAse-free water can be used to elute the RNA sample from the column.
Purification
Various complementary reagents are used in this step to remove contaminants such as polysaccharides, proteins, secondary metabolites, and other possible contaminants. To remove polysaccharides/secondary metabolites and proteins, reagents such as chloroform and isopropyl alcohol are used. Contaminating genomic DNA can also be removed using column-based or enzyme-based methods.
This step purifies and cleans the RNA in the final solution for downstream applications, as well as removes any remaining contaminants. To preserve the extracted RNA, 70% ethanol, water treated with 0.1% (v/v) diethyl pyrocarbonate (DEPC), or RNase-free water can be used. Finally, after dissolving the RNA in one of these reagents, the RNA must be stored at -80°C.
Reverse transcription (RT-qPCR)
There are two methods for quantifying gene expression using RT-qPCR: two-step RT-qPCR and one-step RT-qPCR. RNA is reverse transcribed into cDNA in both cases, and the cDNA is then used as a template for qPCR amplification. The terms one-step and two-step refer to whether RT and real-time PCR amplification are carried out in the same or separate tubes. RNA is first transcribed into cDNA in a reaction using reverse transcriptase in the two-step method. Following that, an aliquot of the resulting cDNA is used as a template for multiple qPCR reactions. The one-step method combines RT and qPCR in the same tube.
Amplification
During the amplification step, the PCR reaction mix, which includes necessary reagents such as DNA polymerase, nucleotides, and primers, is added to the cDNA sample in a PCR tube. The tube is then inserted into a thermal cycler. The temperature cycle is then set to carry out the following steps of the PCR reaction: denaturation, annealing, and extension.
Step : 1 Denaturation
A thermal cycler is used to heat the solution contained in the tube to at least 94°C (201.2°F). The heat breaks the original cDNA sample's hydrogen bonds and separates the cDNA into single strands.
Step : 2 Annealing
The sample mixture is then cooled to between 50°C and 60°C (122 and 140°F), allowing the DNA primers to attach to the individual strands of cDNA that were denatured by heat. This is termed the annealing of the primers.
Step : 3 Extension
The primer attached to the complementary DNA strand is elongated by the DNA polymerase with the help of free nucleotides present in the reaction mix. As a result, double-stranded DNA products are formed. The temperature used during the extension phase is determined by the DNA polymerase.
Generally, the reaction mixture is heated to a temperature intermediate between denaturation and annealing at this stage. 72°C is the ideal temperature for Taq polymerase. The polymerase extends the primers from 5' to 3'.
The length of the target DNA sequence determines the extension time, but a general rule of thumb is one thousand bases per minute at optimal temperature. The amount of target DNA should double during the extension step under ideal conditions.
Analysis
Real-time PCR can be used either quantitatively or semi-quantitatively. It monitors the amplification of a specific DNA molecule during the PCR process rather than at the end, as in conventional PCR.
In the given figure, the number of PCR cycles is shown on the x-axis, and the fluorescence from the amplification reaction is shown on the y-axis, which is proportional to the concentration of amplified product in the tube.
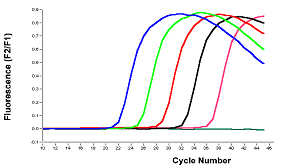
Fluorescence initially remains at background levels despite the fact that the product accumulates exponentially. At some point, enough amplified product accumulates to produce a detectable fluorescence signal. Real-time qPCR can be used to precisely calculate the initial amount of template present in the reaction based on the known exponential function describing the reaction progress because the quantification cycle value is measured in the exponential phase when reagents are not limited.
If there is a large amount of template present at the start of the reaction, only a few amplification cycles are required to accumulate enough product to produce a fluorescence signal above the background level. In contrast, if only a small amount of template is present at the start of the reaction, it will take more amplification cycles to produce a fluorescence signal above the background level.
What is real-time PCR used for?
Real-time quantitative PCR (qPCR) is a simple, low-cost technique used by molecular biologists to measure gene expression. The calculation of a target gene's relative expression by qPCR is based on the use of reference gene(s) as endogenous control(s).
3 more common uses of real-time PCR are given below:
Quantitative analysis of gene expression
Many experimental protocols in scientific research rely on quantitative gene expression analysis. The most widely used method for measuring the level of expression of specific genes is PCR. Gene expression analysis compares the RNA expression levels of multiple genes at the same time. This analysis can help scientists identify differentially expressed genes in diverse pathological states and select gene expression targets for further research.
Diagnosing diseases
qPCR is a widely used laboratory test for the diagnosis of central nervous system (CNS) diseases such as genetic and autoimmune disorders, malignant neoplasms, and infections. With its ability to detect minute amounts of DNA or RNA in tissues or fluids, qPCR has improved diagnostic speed and accuracy, increased understanding of pathogenesis, and helped identify infectious causes for diseases that were previously thought to be idiopathic.
Detecting contaminants
The quantitative polymerase chain reaction (qPCR) method has proven to be an effective tool for detecting and quantifying microorganisms in water. Quantitative PCR assays for the detection of specific adeno- and polyomaviruses, bacteria, and protozoa in various water sources have recently been developed.
Real-time PCR products
Various products can be used in real-time PCR analyses. Some featured products are given below:
Avantor’s real-time PCR (qPCR) products and suppliers
Avantor's internal scientists collaborated with our supplier and research communities to develop solutions for real-time RT-PCR identification of the COVID-19 virus, such as qPCR instruments, consumables, and reagents recommended by leading healthcare organisations. Learn more about Avantor’s qPCR solutions and services here.
AriaMx Real-Time PCR system
The AriaMx Real-Time PCR system is a comprehensive qPCR solution for amplification, detection, and data analysis that combines a novel thermal cycler, an advanced optical system with an LED excitation source, and comprehensive data analysis software.
AriaMx is a closed-tube PCR detection format that can be used with a variety of fluorescence detection chemistries, including SYBR Green and EvaGreen dyes, as well as fluorogenic probe systems like TaqMan. Troubleshooting is aided by functional reporting, over 120 attributes, and built-in diagnostics. Productivity is increased by a modular and flexible design with an intuitive touch-screen interface.
qScript™ 1-Step Virus ToughMix
This is a 2x ready-to-use master mix for one-step or single-tube reverse transcription quantitative PCR detection of RNA viruses such as Flu-A, Flu-B, and SARS-CoV-2 (RT-qPCR). It has been optimised for maximum sensitivity, allowing for the accurate quantification of very small amounts of RNA using dual-labelled hydrolysis probe detection chemistries such as TaqMan® probes in single or multiplexed assay formats. Except for the RNA template and primer/probe assays, qScript 1-Step Virus ToughMix contains all of the components needed for RT-qPCR.